Poster
Impact of 150 mg/kg dosage of paromomycin on gut microbiota in healthy calves
Authors
Pascal Butty Anne Trotel Damien Achard Y. Jaquemet L. Hernandez J. Le Guennec P.-Y. Moalic F. M’Zali
Publication information
European Buiatrics Congress 2025
Objectives
This study aimed to evaluate the effect of oral paromomycin administration over five days on resistance development in the intestinal commensal microbiota of healthy calves.
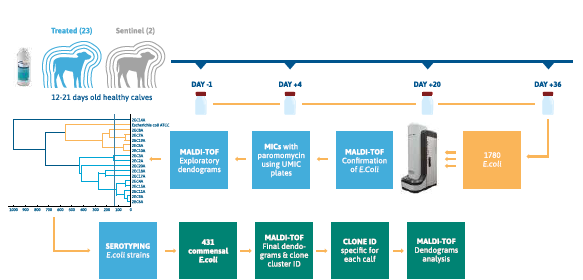
Materials and Methods
Twenty-five healthy calves aged 12 to 21 days, originating from seven French farms, were included in the study. They were housed collectively, fed milk replacers twice daily, and randomized into a treatment group (Gabbrovet Multi®, Ceva Santé Animale, 150 mg/kg daily for five days; n=23) or a control group without treatment (n=2). Daily monitoring included fecal consistency, depression scores, and appetite (evaluated on a 0-2 scale). Fecal samples were collected at four time points: before treatment (Day-1), during treatment (Day+4), and after treatment (Day+20, Day+36). Samples were immediately frozen at -80°C then transferred to the microbiology lab for isolation and microbiological analysis of commensal Escherichia coli strains. For each fecal sample, 20 purified and randomly selected E. coli olonies were selected. To manage the high number of strains, related strains were grouped using mass spectrometry (MaldiTof Biotyper Compass Explorer software). Minimum inhibitory concentrations (MICs) for paromomycin were determined using a customized microdilution method (UMIC), along with aminoglycoside antibiograms following CLSI guidelines. Resistance evolution was tracked by comparing strain data at different times relative to Day-1, using the CA-SFM kanamycin breakpoint for Enterobacteriaceae.
Results
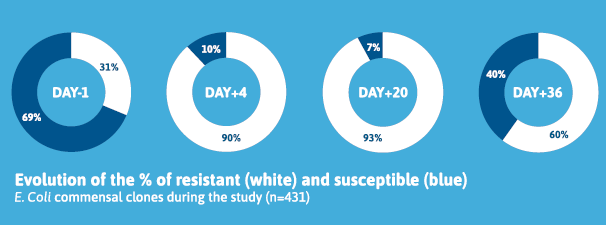
A total of 1,780 E. coli strains were isolated and analyzed. On Day-1, commensal E. coli populations comprised 69% susceptible and 31% resistant clones. Treatment with paromomycin did not induce resistance in previously susceptible E. coli clones. Instead, the treatment eliminated a substantial proportion of susceptible E. coli, resulting in a temporary predominance of preexisting resistant clones within the intestinal microbiota. This disruption was short-lived, as susceptible clones began to reappear post-treatment. Indeed, by Day+36, susceptible E. coli accounted for 40% of the population, compared to 7% on Day+20, 10% on Day+4.
Conclusions
No evidence of resistance acquisition in commensal E. coli was observed during this study. MIC values for individual clones remained stable across sampling points. The paromomycin treatment effectively disrupted the microbiota by favoring preexisting resistant clones, but the flora gradually reverted to its initial composition after treatment cessation. The high bactericidal dosage used in this study likely played a role in preventing resistance development. However, caution is advised for lower dosages (e.g., prophylactic regimen), which are known to increase the likelihood of resistance emergence.
Publication file:




